Dynamics of Membrane Lipids
Page 1 of 1
 Dynamics of Membrane Lipids
Dynamics of Membrane Lipids
Properties of Membranes/Liposomes - Dynamics
Molecules are not static, but rather are dynamic. This also applies to molecular aggregates. In the first part of the section, we will discuss the rigid movement of whole lipid molecules in a bilayer, within a leaftlet and between leaflets. In the second part and the following supplement, we will consider the movement of atoms within a molecule. The movements include motions like bond bending, bond stretching and torsion angle changes like we saw in the previous chapter section on the conformations of n-butane. The position of all atoms within a molecule can be simulated as a function of time - a molecular dynamics simulation. Such motions affect the energy of the molecule, which can be calculated for given atom positions using classical molecular mechanics and electrostatics.
Liposomes and bilayers in general must be somewhat dynamics, otherwise they would be impenetrable barriers across which nothing could pass. Cell membranes must separate the outside of a cell from the inside, but they must also allow passage of molecules and even ions across the membrane. What is the evidence that membranes are dynamic?
First, lipids can diffuse laterally in the membrane. This can be shown as follows. Make a liposome from phosphatidylethanolamine, PE, which has been labeled with TNBS (Trinitrobenzensulfonate). The NH2 on the head group of PE can attach the TNBS which undergoes nucleophilic aromatic substitution with the expulsion of the SO3-2. The TNB group absorbs UV light. Next, using a fluorescent microscope, the fluorescence intensity of a region of the surface can be recorded. Then shine a laser on a small area of the surface, which can photobleach the fluorescence in the area. Over time, fluorescence can be detected from the region again. The rate at which it returns is a measure of the lateral diffusion of the labeled lipids into the region. Lipids can undergo lateral diffusion at a rate of about 2 mm/s. This implies that the lipids can transit the surface of a bacteria in 1 sec.
Transverse, or flip-flop diffusion (movement of a phospholipid from one leaftlet to the other, not within a given leaflet) should be more difficult. Experimentally, this is investigated as shown in the diagrams below.
Liposomes experiments: To test flip-flop diffusion in an artificial membrane, liposomes are made with a mixture of PC and a PC derivative with a nitroxide spin label (has a single unpaired electron). Both inner and outer leaflets of the membrane have the labeled PC. Like a proton in NMR spectroscopy, a single electron has a spin which can give rise to an electron-spin resonance (ESR) signal (as a proton gives rise to a nuclear magnetic resonance signal) when irradiated with the appropriate frequency electromagnetic radiation (microwave frequency for ESR, radio frequency for NMR) in the presence of a magnetic field. The liposomes are kept at 0oC and the ESR signal is determined. Ascorbic acid, a water soluble vitamin and antioxidant/ reducing agent, is added to the liposomes. This reduces the spin labeled PC in the outer leaflet, but not the inner leaftlet of the bilayer since ascorbic acid can not enter the liposome or otherwise interact with it. This reduces the ESR signal to a lower, constant value. The sample is divided into two. One sample is left at 0oC, the other is raised to 30oC. The ESR signal is recorded as a function of time. The 0oC prep shows no change in ESR with time, while the 30oC prep ESR signal decreases with time. This decrease resultsfrom flip-flop diffusion of labeled PC from the inner leaflet to the outer, and subsequent reduction by ascorbic acid. These experiments in experimental bilayer systems like liposomes shows that flip-flop diffusion is orders of magnitude slower than lateral diffusion.
Figure: Flip/Flop diffusion in liposomes A: Making vesicles with ESR active PC analog only in outer leaflet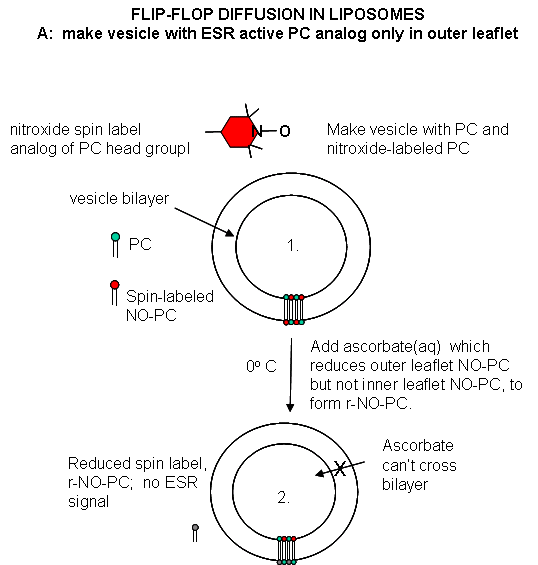 Figure: Flip/Flop diffusion in liposomes B: Raising the temperature to initiate flip/flopdiffusion
Figure: Flip/Flop diffusion in liposomes B: Raising the temperature to initiate flip/flopdiffusion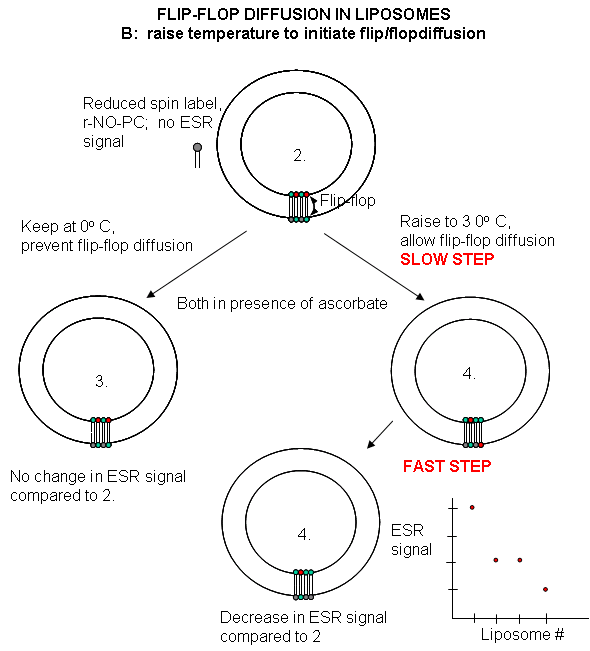
Cell Experiments: An analogous experiment can be done with bacteria. Radiolabeled 32 PO4 is added to cells for one minute, which leads to the labeling of newly synthesized phospholipid (PL) which locates to the inner leaflet. The cells are then split into two samples. One sample is reacted immediately with TNBS, which will label only PE in the outer leaflet. The other sample is incubated 3 minutes (to allow PL synthesis) and then reacted with TNBS. After a short labeling period, the cells are destroyed by adding organic solvents which prevents new lipids biosynthesis. The lipids are extracted into the solvent and then subjected to TLC.
The lipids can be labeled in three ways. Some will be labeled with 32 P alone, some with TNBS alone, and some with both 32P and TNBS. TLC (or other techniques such as HPLC or GC) can easily separate PC and TNBS-labeled PC since they have different structures and hence will migrate to different places on a TLC plate. No chromatographic technique could, however, separate PC and 32 P-PC, since their molecular structure is the same, the only difference being in the nuclei of the P (different number of neutrons).
Those lipids with double labels (TNB and 32 P) must have flipped from the inner leaftlet to the outer leaftlet where they could be labeled with TNBS. The cells incubated for 3 minutes before the addition of TNBS have a much higher level of doubly labeled PL's. Quantitating these data as a function of differing time of incubation at elevated temperatures show that the rate of flip-flop diffusion is much higher in cells than liposomes, which suggests that the process is catalyzed, presumably by a protein transporter (flipase) in cells.
Figure: Flip/Flop diffusion in bacterial cells A: Labeling inner leaflet phospholipids with 32 P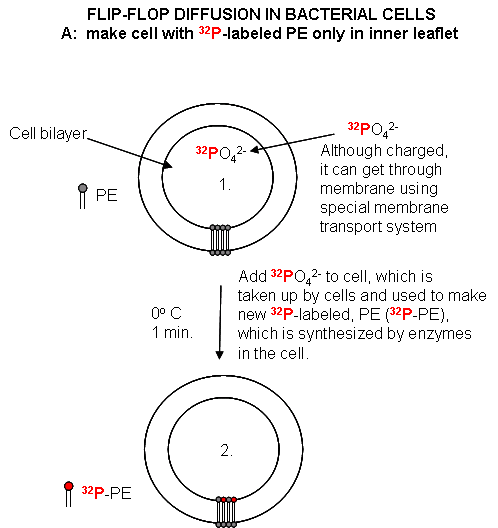 Figure: Flip/Flop diffusion in bacterial cells B: Labeling Cells in A with TNBS to detect Flip/Flop
Figure: Flip/Flop diffusion in bacterial cells B: Labeling Cells in A with TNBS to detect Flip/Flop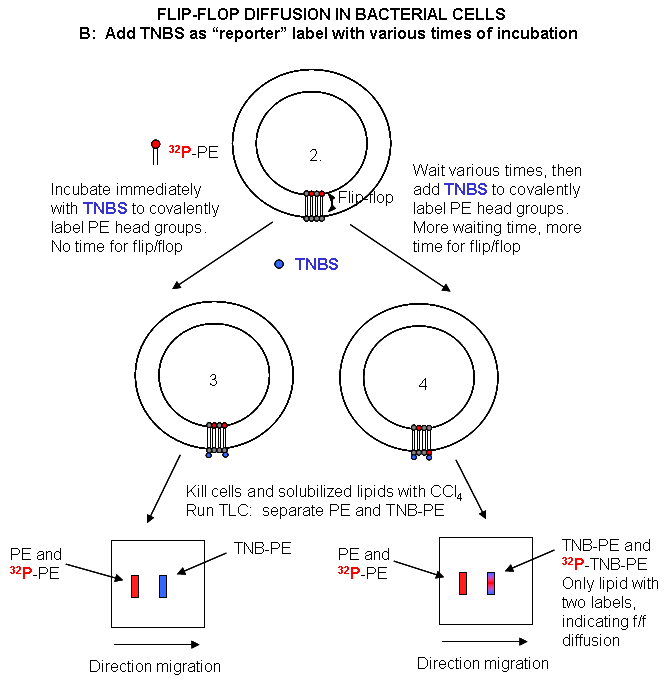
Lipids in membranes are often distributed asymmetrically. The inner and outer leaflet of a biological membrane usually have different PL compositions. For example, in red blood cell membranes, the outer leaflet is composed mostly of sphingomylein (SM) and PC, while the inner leaflet it composed mostly of PE and phosphatidyl serine (PS). This phospholipid contains the amino acid serine linked through its side chain (-CH2OH) to phosphate in position 3 of diacylglyerol. With a negative charge on the phosphate and carboxylate and a positive charge on the amine of PS, this phospholipid is acidic with a net negative charge. All the PS is located in the inner leaftet! This observation will become important latter on, when we discuss programmed cell death. A dying cell will expose PS in the outer leaflet. This is in fact one of the markers of a dying cell.
The membrane lipid composition in an average mammalian cell
| Lipid | % |
| PC | 45-55 |
| PE | 15-25 |
| PI | 10-15 |
| PS | 5-10 |
| PA | 1-2 |
| SM | 5-10 |
| cardiolipin (bis-PG) | 2-5 |
| cholesterol | 10-20 |
 Re: Dynamics of Membrane Lipids
Re: Dynamics of Membrane Lipids
In addition, lipids of a given type often cluster within a leaftlet to form lipid "rafts" which is analogous to a lateral phase separation. Divalent cations like calcium, which can bind to negatively charged PLs like PS, can cause "rafts" of PS to form, giving rise to lateral asymmetry within a leaftlet of a bilayer. Rafts also appear to be enriched in cholesterol and lipids with saturated fatty acids, which would lead to regions of enhanced packing and reduced fluidity. Rafts probably bind or exclude binding of other biological molecules like proteins. Recent studies have shown that the Ebola virus interacts with lipid rafts in the process of entering and exiting the infected cell. Rafts are also involved in how cells sense and respond to their environment. Signaling molecules on the outside of the cell can bind receptor proteins in the membrane. As we will see later, conformational changes in the receptor protein signals the inside of the cells that the receptor is bound with a ligand. Once bound, the receptor can move in the membrane and often cluster in outer leaflet rafts that contain cholesterol and sphingholipids. Inner leaflet rafts have also been observed.
If a liposome preparation is placed in a sensitive calorimeter and the temperature slowly increased, it is observed that the liposome preparation absorbs a significant amount of heat at a temperature characteristic of the PL which compose the liposome. This is analogous to what would happen if solid water was heated. At the melting point of water, an increment of heat is required, the heat of fusion, to break H-bonds and cause melting. Likewise the heat of vaporization is measured when H-bonds are broken on the liquid-gas transition. These transition are associated with non-covalent processes, namely, breaking H-bonds. Graphs of heat absorbed (Q) as a function of temperature, or heat absorbed/T (i.e. the heat capacity) vs temperature are shown below for the melting and evaporation of water, and for liposome transitions.
The transition observed with liposomes is caused by conformational chains in the packing of the acyl chains of the PLs, as the acyl chains change from trans to gauche conformations. These change involve not the simple translation of lipid molecules within and between bilayers but rather the movement of atoms within the molecules. These kinds of motions can be modeled using molecular dynamics simulations.
Before the transition, the acyl chains are more tightly packed in the gel phase, and after they are less tightly packed in the liquid crystalline phase, since many chains are in the gauche conformation.
 Chime Model: Gel and Liquid Crystalline Phases in Phospholipid Bilayers (loads slowly)
Chime Model: Gel and Liquid Crystalline Phases in Phospholipid Bilayers (loads slowly)
The midpoint of the phase transition is called the melting point, Tm. Vesicles in the liquid crystalline phase are more fluid, dynamic, and hence more permeable. Note that the liposomes have not been destroyed but simply have undergone a phase change, much like ice turning to liquid water. Liposomes made of different PL have different Tm.
Vesicles make from PLs with bigger head groups have a lower Tm, since they are less "stable". For example, the Tm for vesicles of di-16:0 versions of PA, PE, and PC have Tm's of 67, 63, and 41 degrees C, respectively. Cholesterol is also a ubiquitous component of animal cell membranes. Its size will allow it to fit into either leaftlet with its polar OH pointed to the outside. One function of cholesterol in membranes is to keep the membrane fluid at any reasonable temperature. When a membrane is at a temperature less than the Tm, it is ordinarily in a gel, not liquid crystalline phase. The cholesterol helps prevent ordered packing of the acyl chains of the PL's, which increases their freedom of motion. Hence the fluidity and permeability of the membrane is increased. At temperatures greater than the Tm, the rigid ring of cholesterol reduces the freedom of the acyl chains to rotation, and hence decreases the number of chains in the gauche conformation. This decreases the fluidity and permeability.
Lipid Conformational Transitions
If a liposome preparation is placed in a sensitive calorimeter and the temperature slowly increased, it is observed that the liposome preparation absorbs a significant amount of heat at a temperature characteristic of the PL which compose the liposome. This is analogous to what would happen if solid water was heated. At the melting point of water, an increment of heat is required, the heat of fusion, to break H-bonds and cause melting. Likewise the heat of vaporization is measured when H-bonds are broken on the liquid-gas transition. These transition are associated with non-covalent processes, namely, breaking H-bonds. Graphs of heat absorbed (Q) as a function of temperature, or heat absorbed/T (i.e. the heat capacity) vs temperature are shown below for the melting and evaporation of water, and for liposome transitions.
Figure: Heat absorption and water: phase transitions 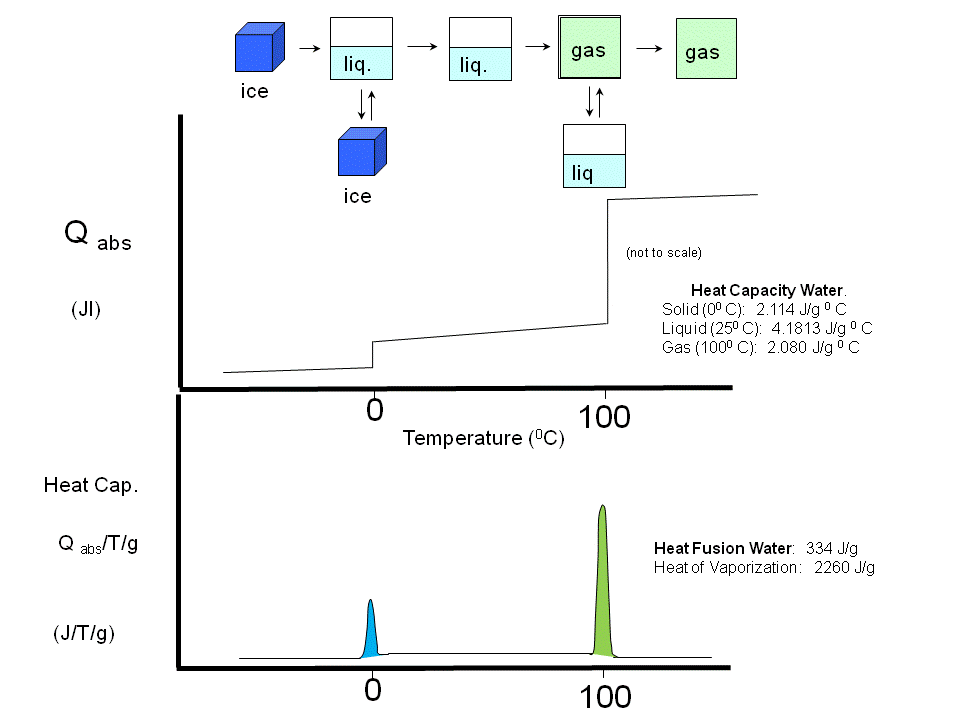

Figure: Differential Scanning Calorimeter (DSC): Phase transition for DPPC (Dipalmitoyl phosphatidylcholine)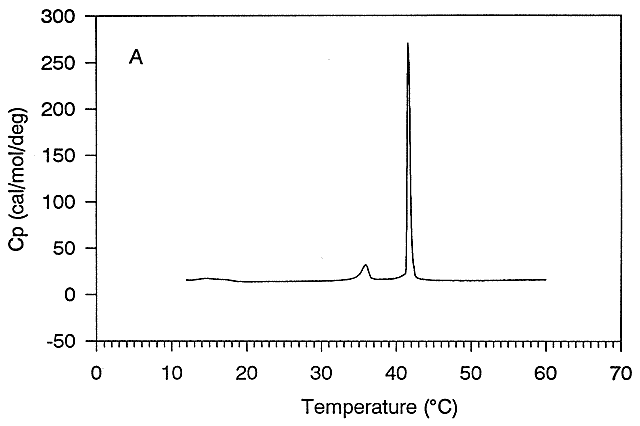

The transition observed with liposomes is caused by conformational chains in the packing of the acyl chains of the PLs, as the acyl chains change from trans to gauche conformations. These change involve not the simple translation of lipid molecules within and between bilayers but rather the movement of atoms within the molecules. These kinds of motions can be modeled using molecular dynamics simulations.
Before the transition, the acyl chains are more tightly packed in the gel phase, and after they are less tightly packed in the liquid crystalline phase, since many chains are in the gauche conformation.
Differences in Gel and Liquid Crystalline Phases in Phospholipids Bilayers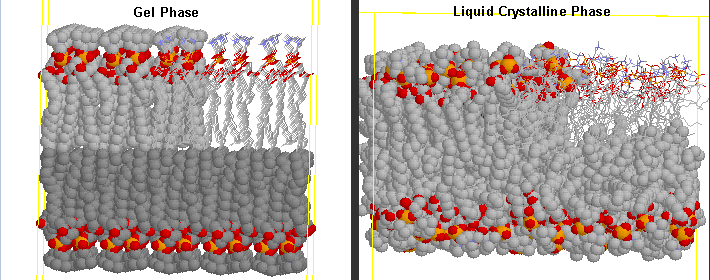

The midpoint of the phase transition is called the melting point, Tm. Vesicles in the liquid crystalline phase are more fluid, dynamic, and hence more permeable. Note that the liposomes have not been destroyed but simply have undergone a phase change, much like ice turning to liquid water. Liposomes made of different PL have different Tm.
Vesicles make from PLs with bigger head groups have a lower Tm, since they are less "stable". For example, the Tm for vesicles of di-16:0 versions of PA, PE, and PC have Tm's of 67, 63, and 41 degrees C, respectively. Cholesterol is also a ubiquitous component of animal cell membranes. Its size will allow it to fit into either leaftlet with its polar OH pointed to the outside. One function of cholesterol in membranes is to keep the membrane fluid at any reasonable temperature. When a membrane is at a temperature less than the Tm, it is ordinarily in a gel, not liquid crystalline phase. The cholesterol helps prevent ordered packing of the acyl chains of the PL's, which increases their freedom of motion. Hence the fluidity and permeability of the membrane is increased. At temperatures greater than the Tm, the rigid ring of cholesterol reduces the freedom of the acyl chains to rotation, and hence decreases the number of chains in the gauche conformation. This decreases the fluidity and permeability.
 Re: Dynamics of Membrane Lipids
Re: Dynamics of Membrane Lipids
Large Scale Conformational Transitions
Large scale conformational transitions can occur in lipid bilayers. These include bud formation, formation of vesicles which split off from the membrane, and fusion of membrane vesicles, all of which are important biologically. To some extent, the tendency to engage in these larger shape transitions depends on the local curvature of the vesicle, which depends on the local phospholipid composition and fluidity of the membrane. We have previously seen that segregation of lipids into rafts (or domains) within a leaflet depends on the phospholipid content. Baumgart et al. have made giant unilamellar vesicles (GUV) containing a mixture of three lipids: sphingomyelin (SM), dioleylphosphatidylcholine (DOPC), and cholesterol (C) to study transitions in the bilayers. SM and C segregate laterally into a "liquid phase" domain characterized by significant order (Lo) while DOPC forms a "liquid phase" with more disorder (Ld). They could detect the different phases through fluorescence microscopy of GUV's labeled with fluorophores (molecules that absorb visible or UV light and emit photons of lower energy (higher wavelength). The first, N-lissamine rhodamine dipalmitoylphosphatidylethanolamine (red fluorescence), partitions almost exclusively into the Ld phase. The second, perylene (blue fluorescence), partitioned with great selectivity into the Lo phase. The results of their studies are shown in the figures below. This images provide stunning visualization of lipid domains (rafts) including those in the shapes of circles, stripes, and rings. Their works shows that the boundary between the domains is important in determining lipid structures. Structures are favored which reduce the perimeter at the boundaries of the phases. It appears that the Ld phase (and associated lipids) is found preferentially in area of high membrane curvature, while the Lo phase is found is regions of lower curvature. The two figures below are reprinted with permission of Nature and the authors: Baumgart, Hess, and Webb, W. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 425, pg 821 (2003) Figure: Equatorial cross sections of GUVs with multiple phases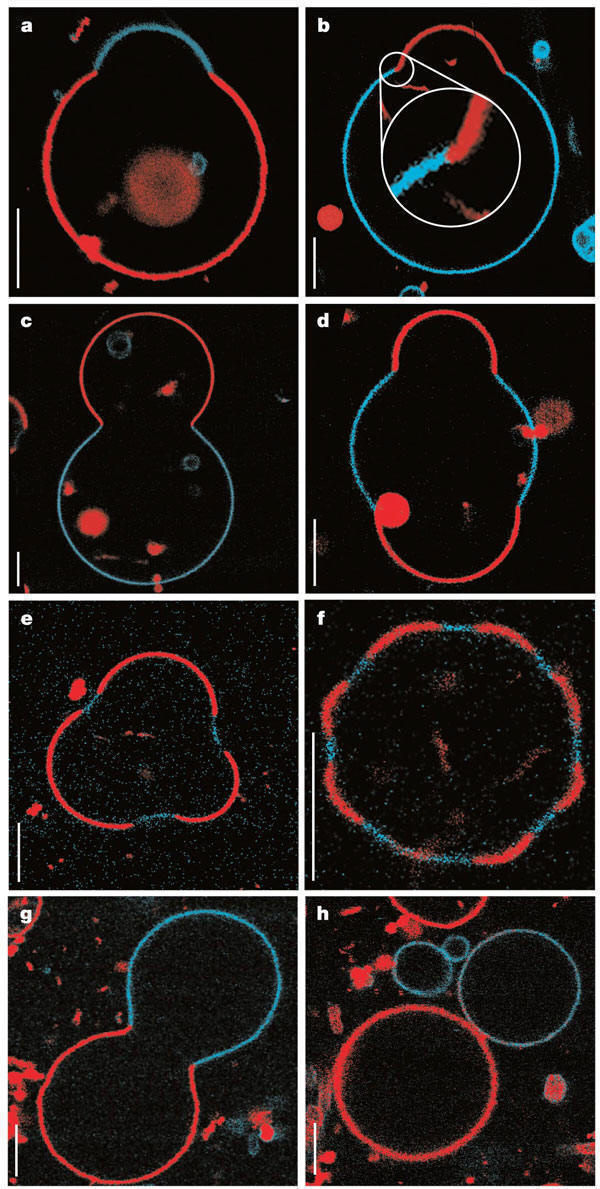 Figure: GUVs structures with multiple phases
Figure: GUVs structures with multiple phases 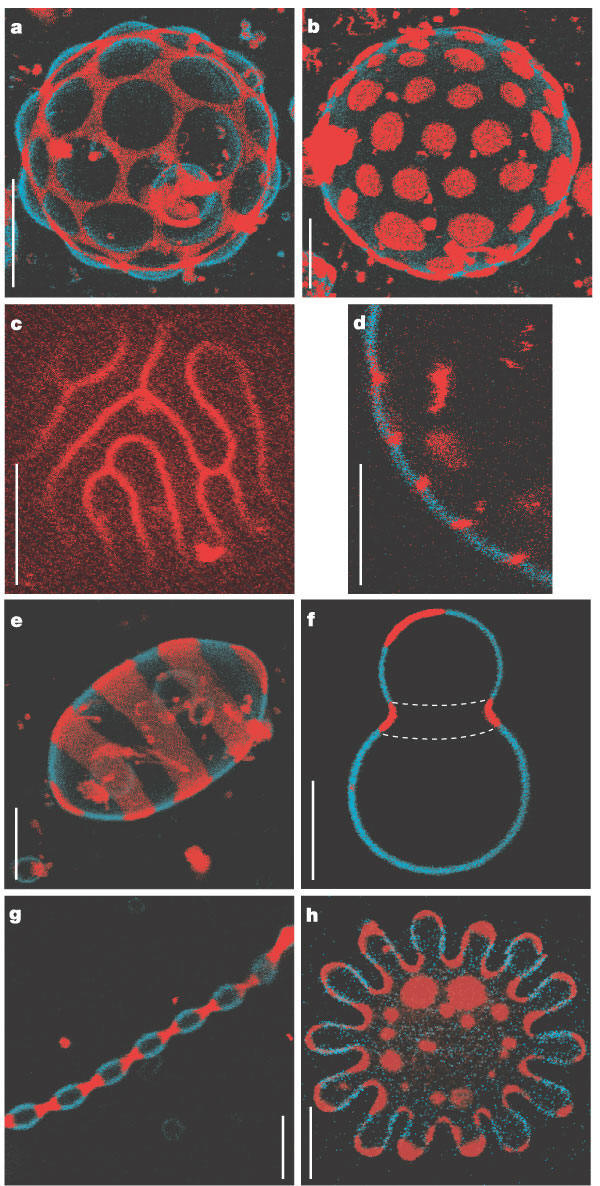
 Figure: GUVs structures with multiple phases
Figure: GUVs structures with multiple phases 
Membrane Permeability
Another indicator of membrane dynamics is the measured permeability coefficient of ions and molecules across the bilayer. (We will discuss these coefficients in the chapter on Passive and Facilitated Diffusion in the Transport and Kinetics Unit.) Permeabilities are correlated to the partition coefficient of the molecule or ion in organic solvents. If the molecule readily dissolves in a nonpolar solvent, it is more likely to pass through the hydrophobic barrier of the membrane. Obviously the size of the molecule would play a part as well. Hence charged ions like Na+ and K+ have low permeabilities. Cl- has a higher permeability, since the charge density on the larger Cl anion is smaller. Water has a surprisingly high permeability although it is polar. It is small and can enter down deep into the head group region through sequential H-bonding which must assist its transfer across the membrane.
Figure: Permeability of liposome bilayers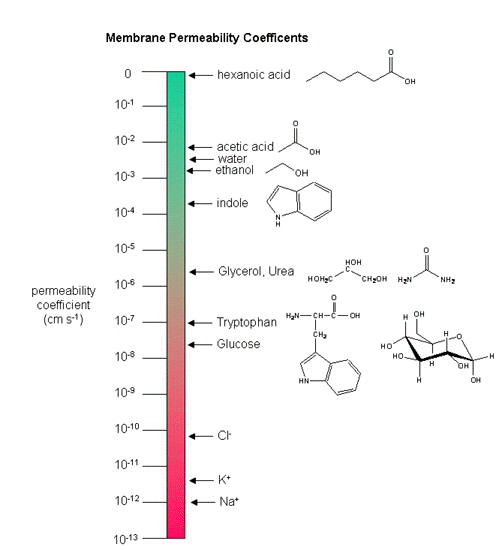

Page 1 of 1
Permissions in this forum:
You cannot reply to topics in this forum|
|
|

